Description
The sequencing analysis service RNA-seq Total Transcriptome from low amount or low quality RNA preparations with Bioinformatics Analyses and Report (Ref-SKU: S.13.1.IT.149.40) based on SMARTer Universal Low Input RNA Kit for Sequencing and ThruPLEX® DNA-Seq Kit provided by Illumina, includes the following for each sample:
- RNA input of any quality and starting from low amount
- Depletion of rRNA or enrichment of polyA+ RNA
- First-strand cDNA synthesis using SMART N6 technology (random primers)
- Amplification of cDNA by PCR
- Removal of SMART adapters
- Library preparation (dsDNA fragmentation, adapters ligation, library amplification)
- Library purification and quantification
- Normalization and Pooling of library
- Sequencing 2×75 PE on NextSeq 550
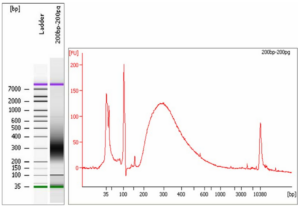
Figure 1: Bioanalyzer analysis of libraries prepared using ThruPLEX DNA-Seq. Libraries were prepared from 200pg DNA (200 bp) using the ThruPLEX DNA-Seq Kit.
Material provided: data
Unit definition: Library preparation: SMARTer Universal Low Input RNA Kit for Sequencing
800 million reads for 2×75 Paired End length (suggested at least 30 million of reads per sample – 24 samples per run). 1 Unit allows testing at least four experimental conditions + two control conditions, with four replicates of each condition = total 24 RNA samples. Four replicates per condition are recommended. Please do not submit projects with less than three replicates per condition, which would be inadequate for statistical analysis.
END-USER RESPONSIBILITIES
Biological material to be provided by the end-users and described in the dedicated request form.
– RNA INPUT RECOMMENDATIONS
Illumina library preparation protocol is followed by the service provider for preparing of the RNA libraries.
The kit protocol is optimized for 200 pg to 10 ng of input RNA.
Minimal sample size to be provided: 50 – 100pg of total RNA per each sample. Minimal volume: 20 ml molecular grade water.
Recommended RNA Integrity Number (RIN) value: < 7. This kit has been validated using input RNA with RIN values between 2 and 3.
– REMARKS:
a) if the provided sample doesn’t comply with the above requested specifications for “RNA INPUT RECOMMENDATIONS”, the customer will be contacted for a re-sampling.
b) in case of failure of the sequencing run, the service will be repeated.
c) to compensate the biological variability within the experiment due to RNA, a biological triplicate for each experimental condition is recommended.
– DESPATCH OF SAMPLES
- Safe-lock 1.5 – 2 ml tubes properly capped to be used to avoid accidental evaporation or sample contamination during shipping.
- Samples to be labelled correctly.
- Correct temperature must be ensured during the shipment to avoid sample degradation.
- Complete the enclosed Sample Submission form and return together with the samples to: customer@pologgb.com.
SUPPLIER RESPONSIBILITIES:
– STORAGE OF BIOLOGICAL SAMPLE
Polo d’Innovazione Genomica Genetica Biologia will store the analysed samples only for 1 month after delivery. Extended storage time is available for a maximum of 4 months and must be requested in advance of the experiments in the service request application.
– STORAGE OF DATA
Polo d’Innovazione Genomica Genetica Biologia waives any responsibility for the storage and backup of the supplied project and sequencing data after delivery to the end-user. Polo d’Innovazione Genomica Genetica Biologia will store the project data only for 1 month after delivery. After expiration of this period, all data are removed from our servers and archived (unless otherwise agreed with the end-user). Your data might be re-imported at an extra cost within the period of 1 year after delivery of the final data. After 1 year, all project data will be deleted permanently from our systems. Extended data storage is available on request at an extra cost and must be requested at project start.
– DELIVERY OF RESULTS
Sequencing results will be sent according to the Modalities of Transfer, with a QC Report PDF File 90 days upon samples delivery by the User.
Modalities of transfer:
- by “WeTransfer” platform for data of up to 2Gb
- by “MegaSync” platform for data ranging from 2Gb to 15Gb
- by HD for data over 15 Gb
Please note that the cost of HD driver is not included. Where requested, the end-user must provide the HD.
For further details on the description of the experiment please consult the kit manuals at the following links:
- SMARTer Universal Low Input RNA Kit for Sequencing User Manual/SMARTer Universal Low Input RNA Kit for Sequencing User Manual
- ThruPLEX DNA-Seq Kit User Manual
For more information, please contact us.
For technical assistance, contact Polo GGB Technical Support:
Email: techsupport@pologgb.com
Telephone: 0039 0577 381310

