Description
Material provided: data
Unit definition: 15 samples – ca 15M reads per sample
Short Description: Detection and characterisation of viruses by metagenomics (VM) is a relatively new technique that takes advantage of the sensitivity of next-generation sequencing (NGS). The characterization of insect viromes has a particular public health significance since mosquitoes and other insect species can transmit human viral pathogens, such as Dengue and Chikungunya viruses. The goals of sample preparation for metagenomic sequencing of viruses are to remove as many nucleic acids as possible from the host and other elements such as bacteria, fungi and parasites, to ensure that most of the virus nucleic acids are retained throughout the process, in order to generate good quality NGS reads. The characterization of viruses based on viral enrichment followed by cDNA sequencing allows the identification of viruses without any prior information about their presence. The Viral Metagenomic sequencing is ideal for the discovery and the profiling of the virome of vector insects.
The experiment and the product
The sequencing analysis service Viral Metagenomic Illumina sequencing (Ref-SKU: S.1.11.IT.149.42), includes the following steps for each sample:
- Reverse-transcription with Maxima H Minus Double-Stranded cDNA Synthesis Kit;
- Nextera XT tagmentation;
- Nextera XT Library Amplification and sample index assignment;
- Library Validation by Qubit and Fragment Analyzer;
- Library Normalization to 4nM for cluster generation;
- Sequencing: 2x 150 PE NextSeq
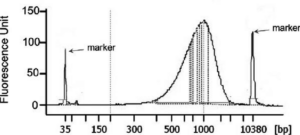
Figure 1: Example of the size distribution of Nextera XT libraries generated from 1 ng of DNA
END-USER RESPONSIBILITIES Biological material to be provided by the end-users and described in the dedicated request form. Any vector is accepted, including vectors resulting from other Infravec2 resources.
– RNA INPUT RECOMMENDATIONS
For the small RNA library preparation, the Polo d’Innovazione Genomica Genetica Biologia, follows the library preparation protocol from Qiagen. The kit protocol has been optimized for Total enriched viral RNA.
Please, provide at least 100ng of total enriched viral RNA. Use fluorometric based method for input RNA quantification. Recommended RNA Integrity Number (RIN) value: ≥ 7
– REMARKS:
- If the provided sample doesn’t comply with the above requested specifications for “RNA INPUT RECOMMENDATIONS”, the customer will be contacted for a re-sampling.
- In case of failure of the sequencing run, the service will be repeated.
– DESPATCH OF SAMPLES
- Safe-lock 1.5 – 2 ml tubes properly capped to be used to avoid accidental evaporation or sample contamination during shipping.
- Samples to be labelled correctly.
- Correct temperature must be ensured during the shipment to avoid sample degradation.
- Complete the enclosed Sample Submission form and return together with the samples to: orders@pologgb.com.
SUPPLIER RESPONSIBILITIES:
– STORAGE OF BIOLOGICAL SAMPLE
Polo d’Innovazione Genomica Genetica Biologia will store the analysed samples only for 1 month after delivery. Extended storage time is available for a maximum of 4 months and must be requested in advance of the experiments in the service request application.
– STORAGE OF DATA
Polo d’Innovazione Genomica Genetica Biologia waives any responsibility for the storage and backup of the supplied project and sequencing data after delivery to the end-user. Polo d’Innovazione Genomica Genetica Biologia will store the project data only for 1 month after delivery. After expiration of this period, all data are removed from our servers and archived (unless otherwise agreed with the end-user). Your data might be re-imported at an extra cost within the period of 1 year after delivery of the final data. After 1 year, all project data will be deleted permanently from our systems. Extended data storage is available on request at an extra cost and must be requested at project start.
– DELIVERY OF RESULTS
The sequencing results will be sent according to the Modalities of Transfer, with a QC Report PDF File 90 days upon samples delivery by the User.
Modalities of transfer:
- by “WeTransfer” platform to email for data of up to 2Gb
- by “MegaSync” platform for data ranging from 2Gb to 15Gb
- by HD for data over 15 Gb
Please note that the cost of HD driver is not included. Where requested, the end-user must provide the HD.
Description of the experiment and product:
For further details on the description of the experiment please consult: “Nextera XT library preparation kit Guide”; “Maxima H Minus Double-Stranded cDNA Synthesis Kit guide”
For more information, please contact us.
For technical assistance, contact Polo GGB Technical Support:
Email: techsupport@pologgb.com
Telephone: 0039 0577 381310

